Welcome to pCysMod
Protein cysteine modifications mainly include thiol oxidation and palmitoylation of thioesterification reaction, which lead to a cascade of biochemical reactions. There are many types of cysteine modifications mediated by thiol groups, such as S-nitrosylation, S-sulfhydration, S-sulfenylation and S-sulfinylation. Besides, S-palmitoylation is cysteine modification through thioesterification reaction. These modifications lead to a cascade of biochemical reactions, and regulate various physiological and pathological processes, such as autophagy, redox homeostasis and cell signaling, demonstrating a closely relationship with many human diseases including cancers and diabetes and so on.
In this work, we constructed pCysMod to predict multiple protein cysteine modification server. The predictor is developed based on deep learning. Three bright spots of pCysMod software are summarized below: i) this is a comprehensive prediction tool for cysteine modifications, including S-nitrosylation, S-palmitoylation, S-sulfhydration, S-sulfenylation and S-sulfinylation; ii) four kinds of protein sequence features are modeled including binary encoding profiles (BE), amino acid composition (AAC), position-specific scoring matrix (PSSM), composition of k-spaced amino acid pairs (CKSAAP); iii) to facilitate further analysis, three fundamental properties of protein sequence are visualized in the result page, including disorder, surface accessibility and secondary structure.
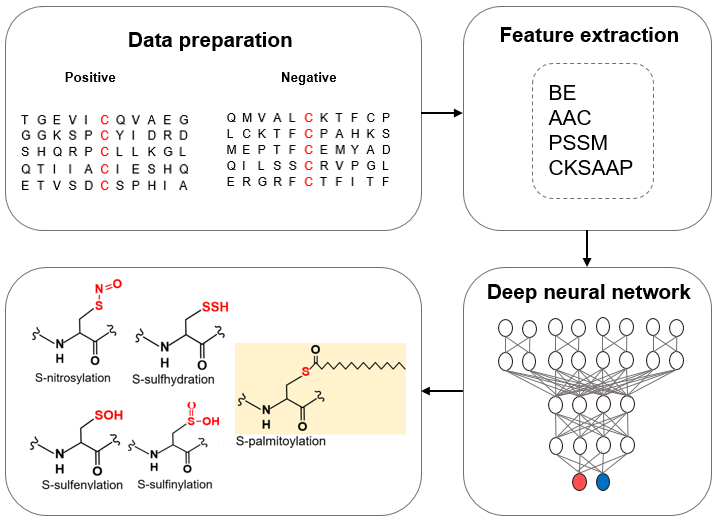
Deep neural network for prediction of multiple cysteine modifications
• Four sequence features of the cysteine modifications sites are extracted to build models.
• Deep-learning method is used to predict cysteine modifications.
• Model of pCysMod is a Comprehensive prediction tool for cysteine modifications, including S-nitrosylation, S-palmitoylation, S-sulfhydration, S-sulfenylation and S-sulfinylation.
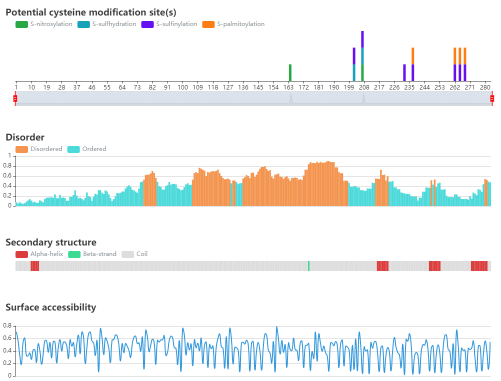
Protein sequence and structure properties
• The potential cysteine modification sites of the query sequence are indicated.
• IUPred is used to predict the disorder region.
• NetSurfP is used to predict the surface accessibility and secondary structure.
